Goal
The overarching goal of the project was threefold. First, we integrated digital imaging into Temple University Hospital’s clinical operations. Then, we established a public database of pathology slides. Lastly, we developed an image classification system to automatically annotate each pathology slide. The result was a fully functional digital pathology imaging system. After the database of pathology slides were annotated using this system, they were archived and used for support, education, and/or research purposes.
Hardware
 Pathology slides are scanned with the Leica Aperio AT2 Digital Whole
Slide Scanner (DWSS). The Aperio AT2 has a 400-slide capacity
brightfield scanner allowing a high throughput rate of approximately
50 slides per hour at 20x magnification. The scanning software uses
lossless compression and generates high quality TIFF files ranging in
size from 250 megabytes to 5 gigabytes (resolution dependent).
Pathology slides are scanned with the Leica Aperio AT2 Digital Whole
Slide Scanner (DWSS). The Aperio AT2 has a 400-slide capacity
brightfield scanner allowing a high throughput rate of approximately
50 slides per hour at 20x magnification. The scanning software uses
lossless compression and generates high quality TIFF files ranging in
size from 250 megabytes to 5 gigabytes (resolution dependent).
 Our App Server is what hosts the Leica Aperio eSlide Manager and
Image Scope applications. The eSlide Manager is a web based
application that pathologists can use to view digitized pathology
slides and export specimen data to Image Scope; which is simply
a local application.
Our App Server is what hosts the Leica Aperio eSlide Manager and
Image Scope applications. The eSlide Manager is a web based
application that pathologists can use to view digitized pathology
slides and export specimen data to Image Scope; which is simply
a local application.
 To store the slides, we used a cost-effective Petabyte server. There
are two Petabyte machines in total, one to store the digital slides,
and one as backup. Since the digital pathology slides we use can sometimes
have a size of up to 5 gigabytes each, we needed as much storage as we
could get. Our Petabyte machine collected a 10-year digitized archive
of digital pathology slides. Each Petabyte machine contains two JBODs
with each containing 60 hard drives each. This means that there are
240 total hard drives installed on both machines combined.
To store the slides, we used a cost-effective Petabyte server. There
are two Petabyte machines in total, one to store the digital slides,
and one as backup. Since the digital pathology slides we use can sometimes
have a size of up to 5 gigabytes each, we needed as much storage as we
could get. Our Petabyte machine collected a 10-year digitized archive
of digital pathology slides. Each Petabyte machine contains two JBODs
with each containing 60 hard drives each. This means that there are
240 total hard drives installed on both machines combined.
Data Development
Our department has a successful history in managing medical databases. The TUH EEG Corpus now exceeds over 50,000 EEGs spanning the years 2002 to present. For this project, we have digitized a 10-year archive of existing slides and integrated digitization into ongoing clinical operations. Our student researchers worked side-by-side with clinical and research pathologists to develop best practices for system optimization. After a baseline system was established, the data was released for review. We collected feedback from users with respect to the scan parameters and organization of the public data to maximize the value of the data for the community.
Algorithm Development
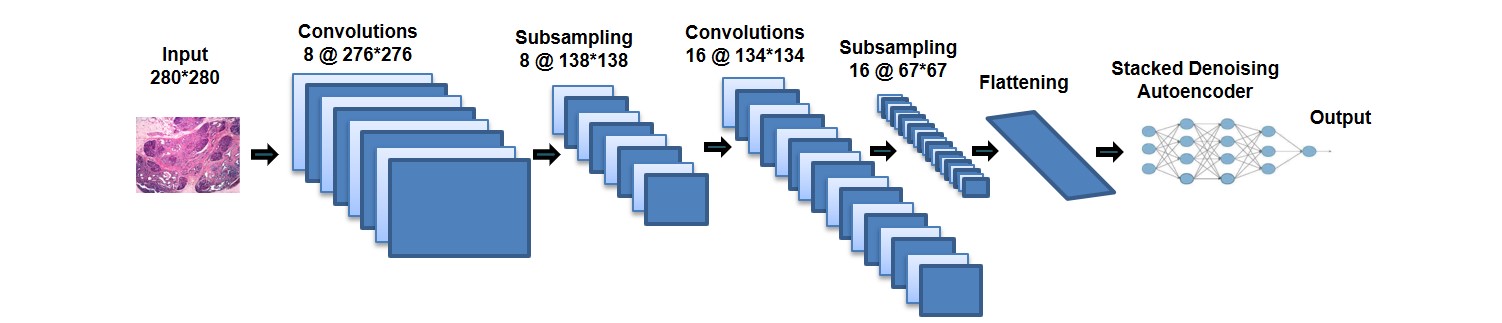 We adapted our existing TUH EEG Corpus deep learning system to
this project. Once a baseline system was in place, we compared
our analysis with physician feedback and adjusted the system
accordingly. The software was developed according to strict
design and formatting requirements and included regression
testing and built-in certifications. Our software was developed
using Python and executes on general purpose processors and high-speed
GPUs. As always, the data and software we developed is open-source.
We adapted our existing TUH EEG Corpus deep learning system to
this project. Once a baseline system was in place, we compared
our analysis with physician feedback and adjusted the system
accordingly. The software was developed according to strict
design and formatting requirements and included regression
testing and built-in certifications. Our software was developed
using Python and executes on general purpose processors and high-speed
GPUs. As always, the data and software we developed is open-source.
 NSF MRI: High Performance Digital Pathology Using
Big Data and Machine Learning
NSF MRI: High Performance Digital Pathology Using
Big Data and Machine Learning
